ಟ್ವೀಟ್ಗಳು
- ಟ್ವೀಟ್ಗಳು, ಪ್ರಸ್ತುತ ಪುಟ.
- ಟ್ವೀಟ್ಗಳು & ಪ್ರತಿಕ್ರಿಯೆಗಳು
- ಮಾಧ್ಯಮ
ನೀವು @Eric_Fauman ಅವರನ್ನು ತಡೆಹಿಡಿದಿರುವಿರಿ
ಈ ಟ್ವೀಟ್ಗಳನ್ನು ವೀಕ್ಷಿಸಲು ನೀವು ಖಚಿತವಾಗಿ ಬಯಸುವಿರಾ? ಟ್ವೀಟ್ ವೀಕ್ಷಣೆಯು @Eric_Fauman ಅವರ ತಡೆತೆರವುಗೊಳಿಸುವುದಿಲ್ಲ
-
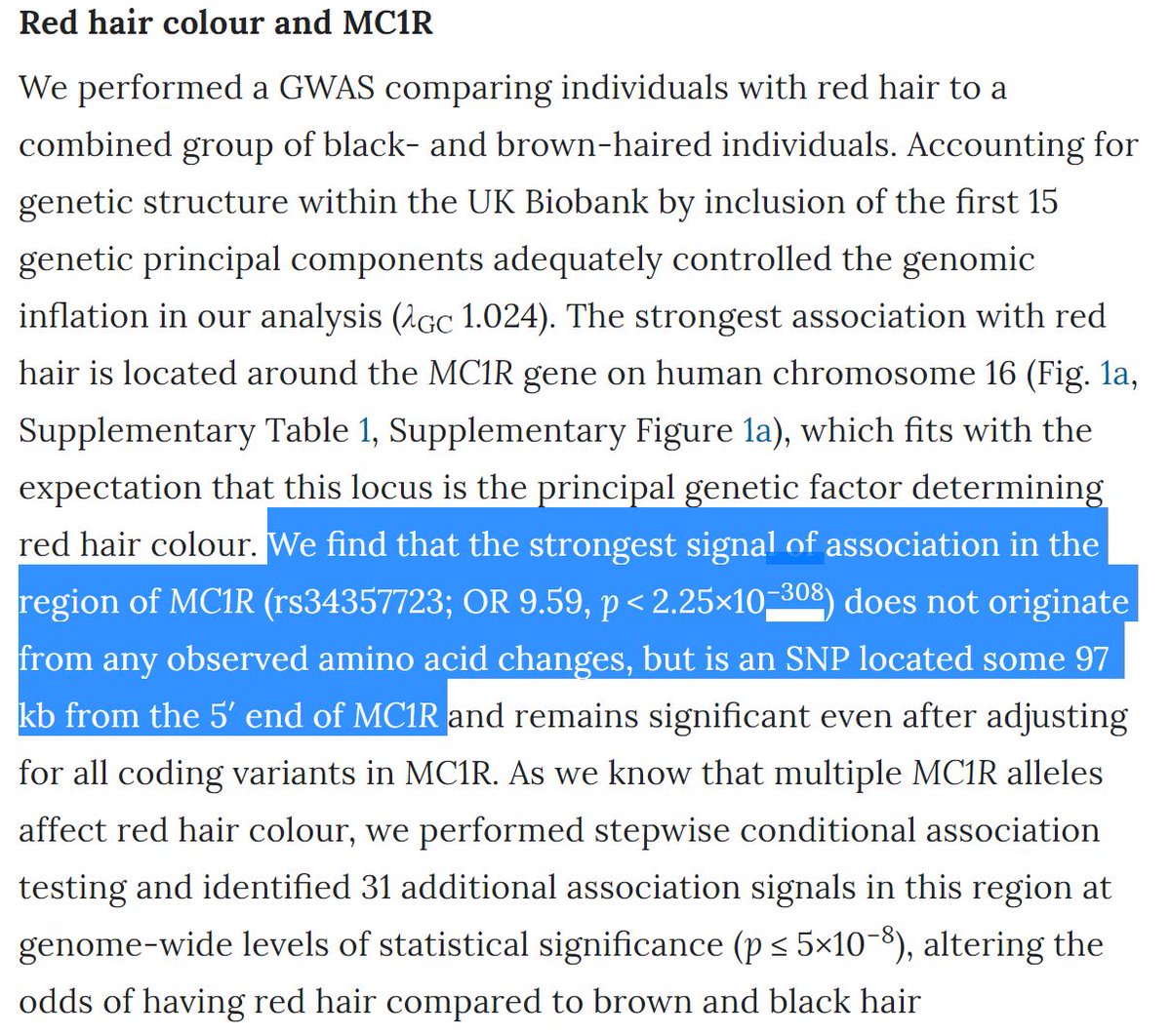
Of course p-values this extreme lose statistical interpretation but more of our GWAS will be leading to p-values of this magnitude. Excel, R and other common applications have issues with numbers like these, but we need to be prepared to appropriately handle these associations.
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
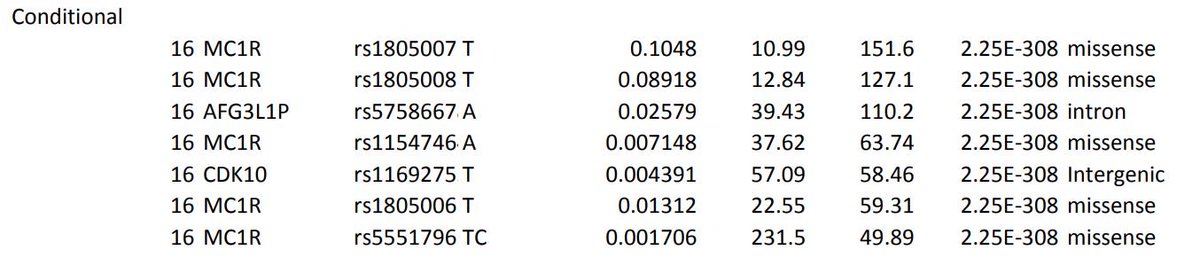
Why is this an issue? I think it affects the interpretation of the lead SNPs. I think this sentence from the paper is incorrect as it looks like the coding variant *is* the most significant at MC1R. All the MC1R SNPs were assigned the same p-value even though the zstats differpic.twitter.com/exiLSKLi39
 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Why didn't this come out in the recent
@uk_biobank#GWAS on red hair color published last week: https://www.nature.com/articles/s41467-018-07691-z …@MDMorgan_cam? I think it's because of the underflow issue that capped all extreme p-values arbitrarily to 2.25e-308ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
The most extreme p-value I've found is 4e-9952. Over 3 orders of magnitude _of orders of magnitude_ more extreme than our standard GWAS cutoff of 5e-8. The trait is "red hair" and the SNP is the R151C missense mutation in MC1R https://www.snpedia.com/index.php/Rs1805007 …
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
If you're not a fan of extreme p-values in
#GWAS you can stop reading right here. But since I've been following the@uk_biobank Manhattan plots generated by@GWASbot from the@bmneale group I thought I'd "skip to the end" and look up most extreme p-value for any trait/any SNP ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
As a Pfizer scientist I work alongside talented immigrants from all over the world.
@Pfizer - why are you advertising on Tucker Carlson? Do you support his views on immigrants? What message are you sending to our colleagues and customers?https://twitter.com/thehill/status/1073704709465223168 … ಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
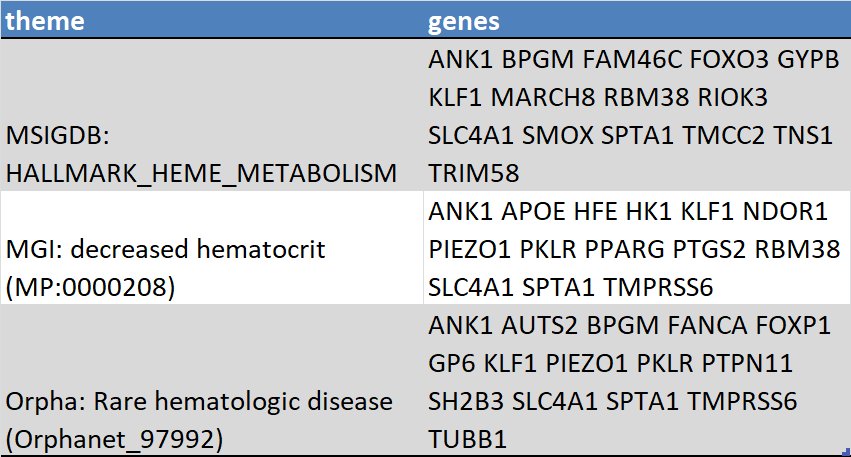
In a robust GWAS the underlying biology comes screaming through. Simply looking at the closest gene to each lead SNP turns up good enrichment for relevant pathways:pic.twitter.com/lHbqgarB40
 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
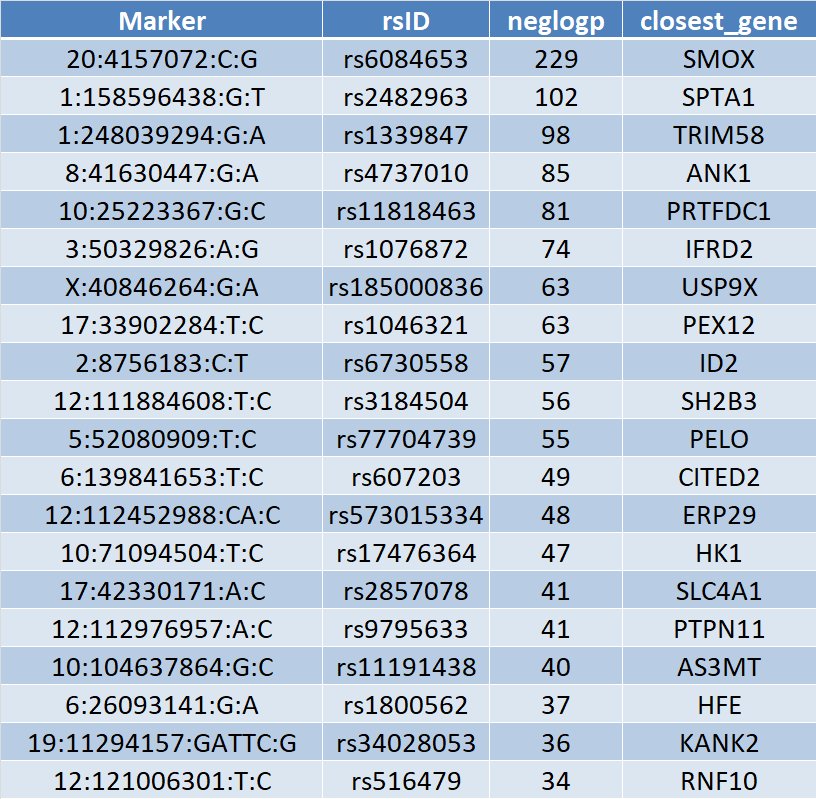
A crude distance cutoff (500kb) yields 270 lead SNPs with p < 5e-8. Here are the top 20 loci, simply annotated with the closest gene:pic.twitter.com/xXEHdHrjIj
 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
A nice example of what's possible in an automated analysis of the UKBB data, but the definitive reference for this trait is from the meta-analysis of UKBB+Interval by
@aidanbutty,@nicolesoranzo and colleagues https://www.ncbi.nlm.nih.gov/pubmed/?term=27863252 …https://twitter.com/SbotGwa/status/1072557472454467586 …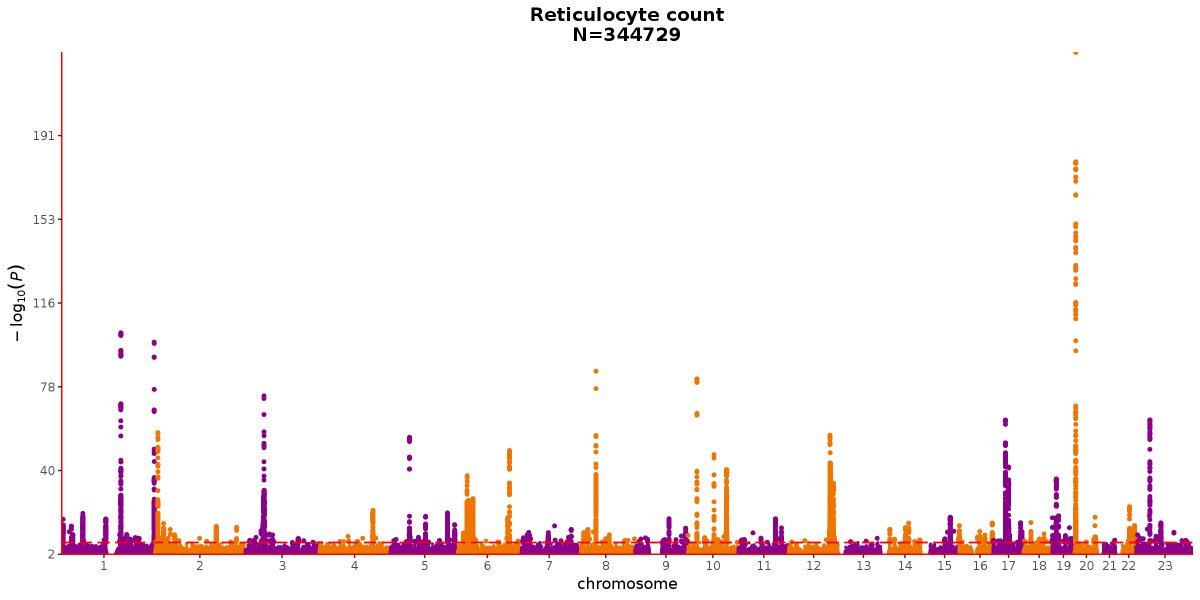 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
4 of the 5 hits are also hits in the bone mineral density GWAS. The last, leptin receptor, may have a role in bone repair.
@BrentRichards19 - any thoughts?pic.twitter.com/LkYsVVElks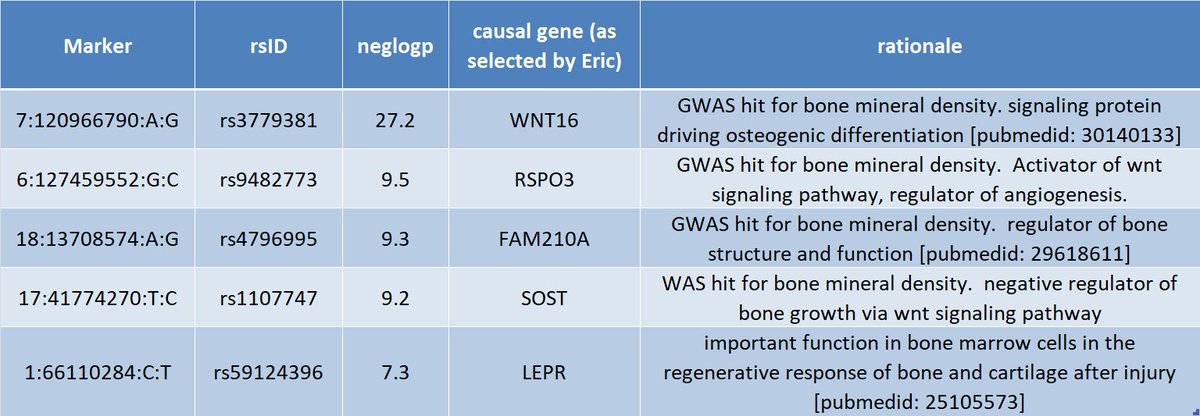 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Knee Arthrosis, Hip Arthrosis and Wrist Fracture - could there be a connection? Spoiler alert, no overlapping signals, but all 5 hits for this GWAS by a biological interpretationhttps://twitter.com/SbotGwa/status/1070745529435459584 …
 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Interestingly, no apparent overlap with the hits from last week's GWAS on knee arthrosis. I don't know enough about the etiology of either disease to know if this makes sense. Hits today tend to focus more on limb development and less on bone mineral density.
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
6 of the top hits for the GWAS of hip arthrosis implicate genes known to be causal for rare diseases involving bone development (according to
@Orphanetpic.twitter.com/nzm4ArQV8m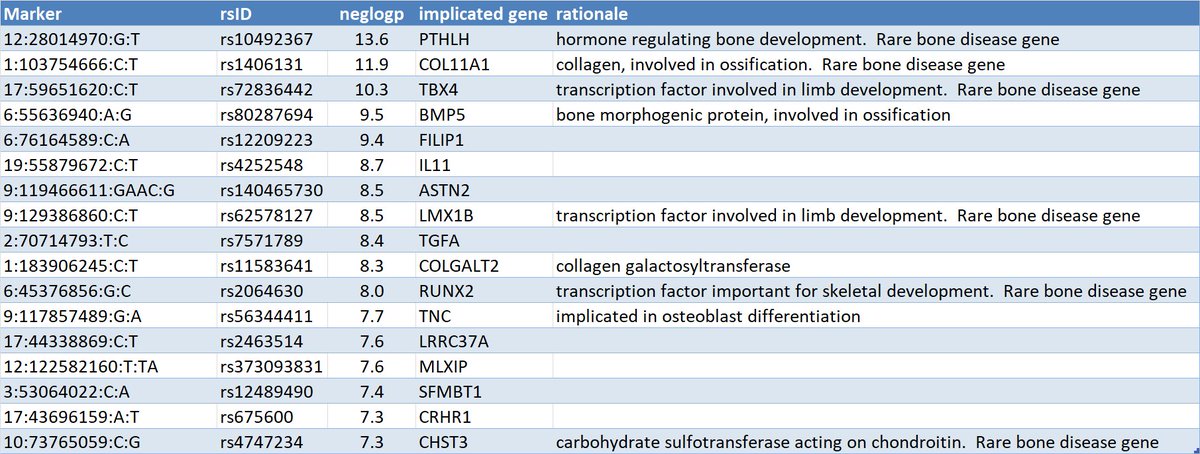 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Good overlap with known bone development genes. Undoubtedly much new biology to be learned here as well:
@uk_biobankhttps://twitter.com/SbotGwa/status/1070383142412959744 …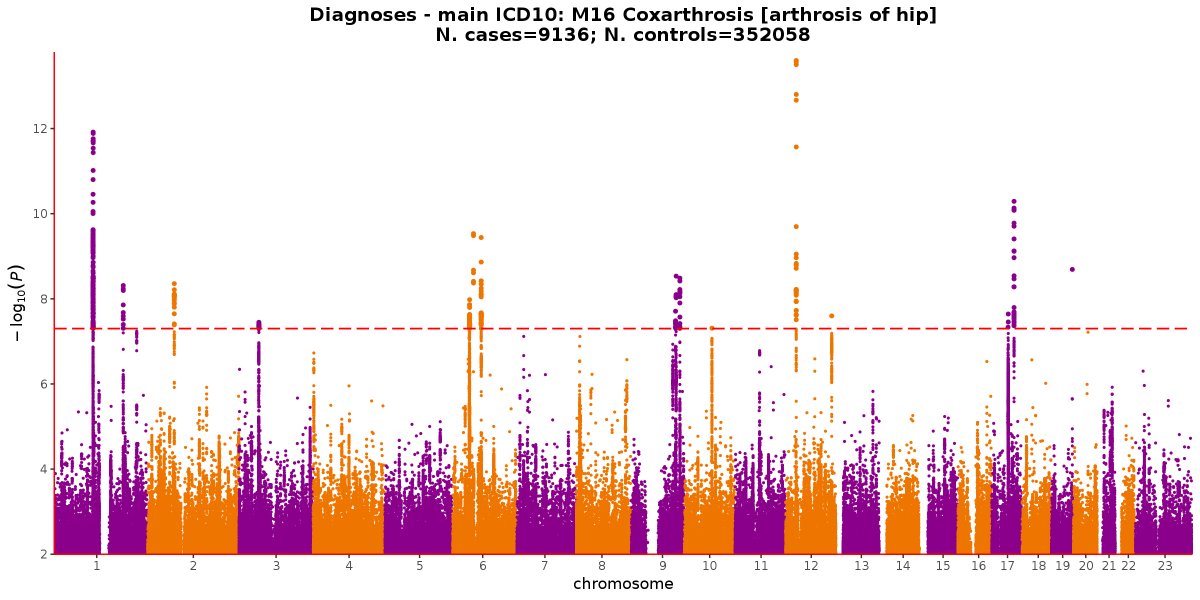 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
And one of my favorite figures that links pretty much all the loci implicated by this GWASpic.twitter.com/zFmJc0gZHM
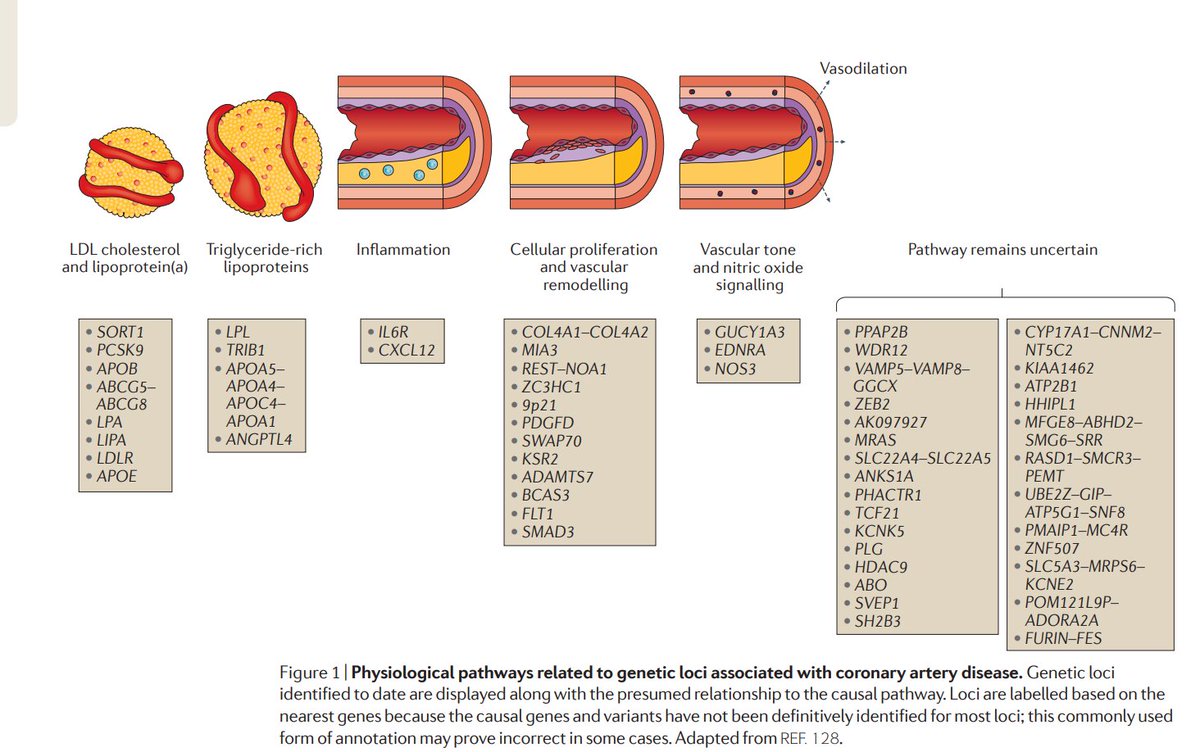 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Basically all these loci are very well explained in this excellent review by my favorite twitterer,
@skathire Genetics of coronary artery disease: discovery, biology and clinical translation: http://www.kathiresanlab.org/our-publications/genetics-coronary-artery-disease-discovery-biology-clinical-translation/ … First author:@amitvkheraಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
Basically looks like a solid GWAS on cholesterol levels with maybe some T2D risk thrown in as well:pic.twitter.com/drZC2UGkv1
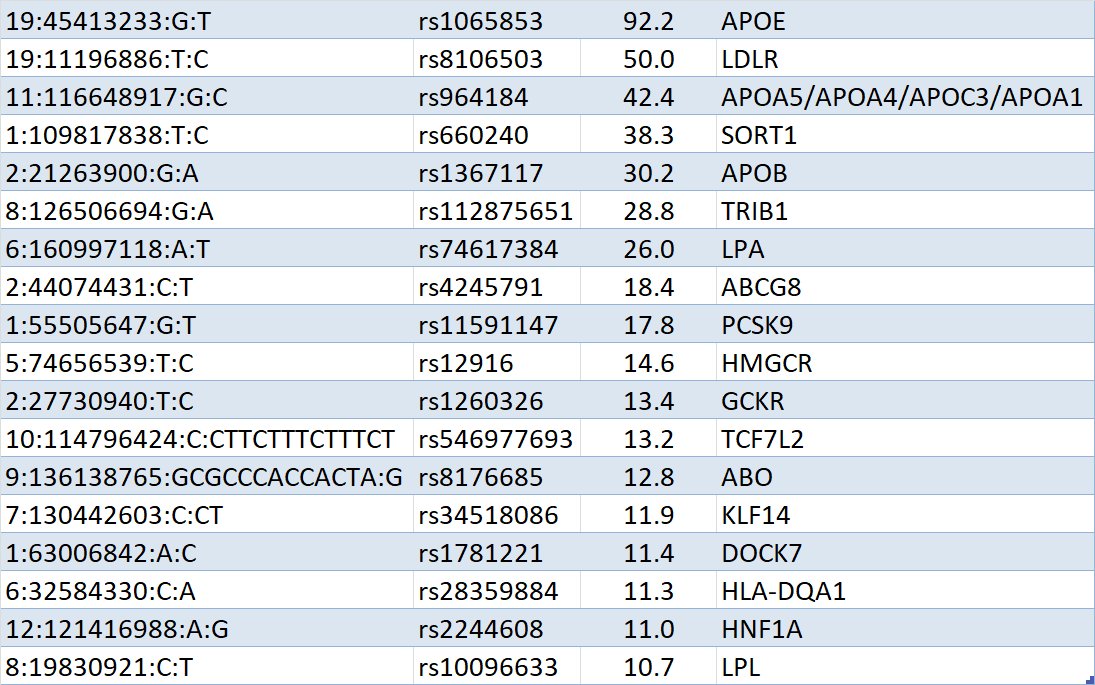 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
In which we find genetic support for 2 current cholesterol lowering therapieshttps://twitter.com/SbotGwa/status/1070020752236310528 …
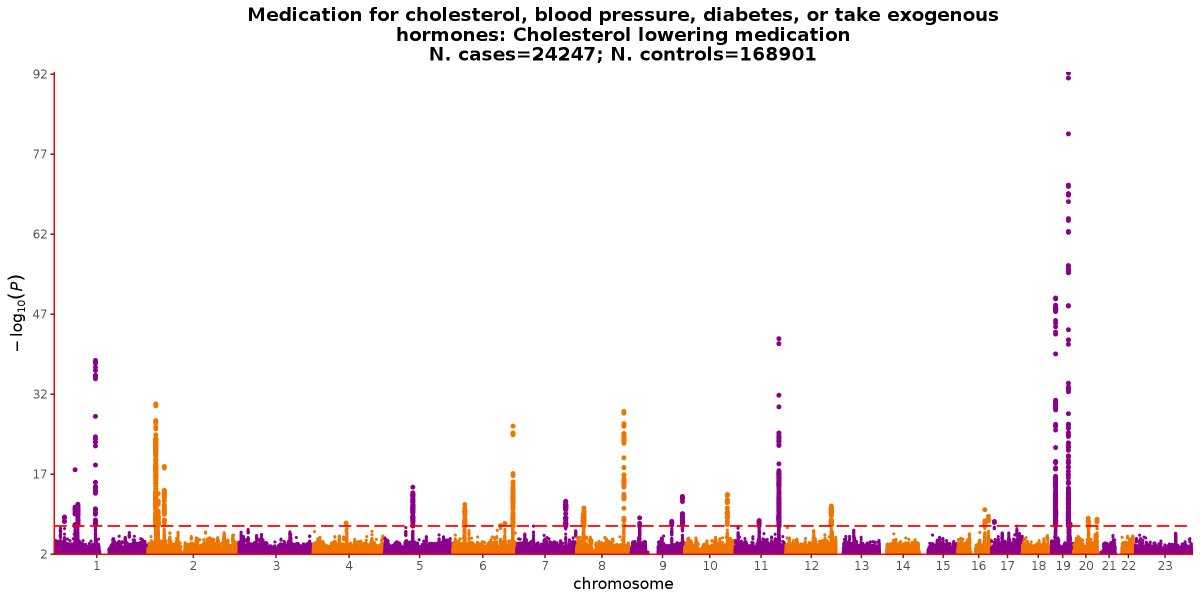 ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
So this GWAS illustrates two themes I've gleaned from reviewing all of
@SbotGwa's output: 1) The closest gene is most often the correct causal gene, and 2) hit weaker than about 1e-10 are suspect because of the sheer number of traits considered. Thoughts?ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು -
The 5th hit is barely above genome-wide significance and sits in the gene CPS1. CPS1 has come up in numerous GWAS for metabolic traits (e.g., glycine) but this SNP is not in LD with those SNPs. My best guess is that the hit here is spurious.
ಈ ಥ್ರೆಡ್ ತೋರಿಸಿಧನ್ಯವಾದಗಳು. Twitter ಇದನ್ನು ನಿಮ್ಮ ಕಾಲರೇಖೆಯನ್ನು ಉತ್ತಮಗೊಳಿಸಲು ಬಳಸುತ್ತದೆ. ರದ್ದುಗೊಳಿಸುರದ್ದುಗೊಳಿಸು
ಲೋಡಿಂಗ್ ಸಮಯ ಸ್ವಲ್ಪ ತೆಗೆದುಕೊಳ್ಳುತ್ತಿರುವಂತೆನಿಸುತ್ತದೆ.
Twitter ಸಾಮರ್ಥ್ಯ ಮೀರಿರಬಹುದು ಅಥವಾ ಕ್ಷಣಿಕವಾದ ತೊಂದರೆಯನ್ನು ಅನುಭವಿಸುತ್ತಿರಬಹುದು. ಮತ್ತೆ ಪ್ರಯತ್ನಿಸಿ ಅಥವಾ ಹೆಚ್ಚಿನ ಮಾಹಿತಿಗೆ Twitter ಸ್ಥಿತಿಗೆ ಭೇಟಿ ನೀಡಿ.


